|
chr20_-_34243118
|
42.454
|
|
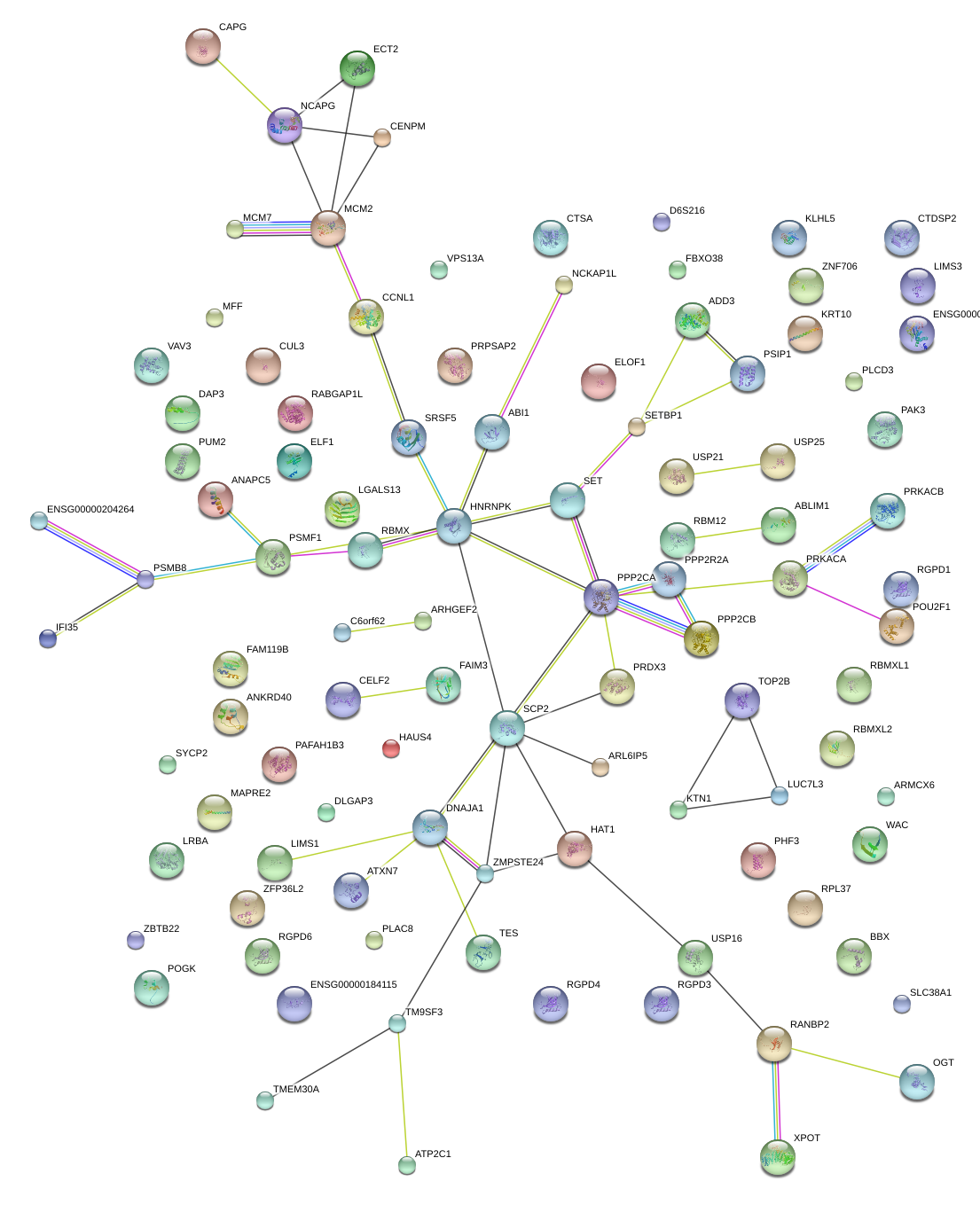
RBM12
|
RNA binding motif protein 12
|
|
chr10_+_28878666
|
31.705
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr10_+_11047253
|
30.233
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_172821874
|
29.669
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr17_+_18768781
|
28.741
|
NM_001243940
|
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2
|
|
chr21_+_30400229
|
26.371
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr17_+_48821106
|
21.488
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr10_+_11047274
|
20.354
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr9_-_86584079
|
20.035
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr2_+_109204904
|
19.224
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr1_+_167298280
|
16.545
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr11_-_67450536
|
16.292
|
|
RPL37
|
ribosomal protein L37
|
|
chr10_-_27057776
|
15.893
|
|
ABI1
|
abl-interactor 1
|
|
chr3_+_63884074
|
15.296
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr12_-_46633551
|
15.234
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr1_+_84630575
|
14.996
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr13_-_41593405
|
14.946
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr1_+_40735749
|
14.432
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr2_-_225370687
|
14.166
|
|
CUL3
|
cullin 3
|
|
chr9_+_79968344
|
13.864
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr3_-_25683825
|
13.655
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr2_-_20527120
|
12.794
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chrX_+_70765479
|
12.384
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr2_-_20512016
|
12.332
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr2_+_109204777
|
11.922
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr1_-_207095198
|
11.630
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr19_-_42806928
|
11.625
|
NM_001145939
NM_001145940
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa)
|
|
chr4_-_84035888
|
11.541
|
|
PLAC8
|
placenta-specific 8
|
|
chr14_+_70236266
|
11.455
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr10_+_111765725
|
11.305
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr6_-_32812711
|
11.142
|
NM_004159
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7)
|
|
chr3_+_63953419
|
11.034
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr13_-_41593491
|
10.515
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr4_-_84035908
|
10.446
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr17_+_41158741
|
10.393
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr9_+_33026482
|
10.331
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chrX_-_135962872
|
10.182
|
|
RBMX
|
RNA binding motif protein, X-linked
|
|
chr6_-_24719286
|
10.178
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr10_-_120938337
|
10.158
|
NM_006793
NM_014098
|
PRDX3
|
peroxiredoxin 3
|
|
chr12_+_64803741
|
10.136
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr5_-_168271852
|
9.926
|
|
|
|
|
chr1_-_108231122
|
9.922
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr2_-_43453737
|
9.781
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr8_+_26150731
|
9.742
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chrX_-_135962929
|
9.713
|
NM_001164803
NM_002139
|
RBMX
|
RNA binding motif protein, X-linked
|
|
chr1_+_53480573
|
9.523
|
NM_001007099
NM_001007100
NM_001007250
|
SCP2
|
sterol carrier protein 2
|
|
chr3_-_156878481
|
9.494
|
NM_020307
|
CCNL1
|
cyclin L1
|
|
chr1_+_53480616
|
9.299
|
|
SCP2
|
sterol carrier protein 2
|
|
chr10_-_116392099
|
9.170
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr17_+_26794838
|
9.141
|
|
|
|
|
chr4_-_151770611
|
9.124
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr20_+_44518782
|
8.764
|
|
CTSA
|
cathepsin A
|
|
chr6_-_75970566
|
8.756
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr22_+_31366662
|
8.745
|
|
|
|
|
chr2_+_109204745
|
8.652
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_-_43453146
|
8.444
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr14_-_23426356
|
8.408
|
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr10_-_120938276
|
8.376
|
|
PRDX3
|
peroxiredoxin 3
|
|
chr1_-_147931877
|
8.294
|
|
FLJ39739
|
uncharacterized FLJ39739
|
|
chr14_+_56078735
|
8.265
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr1_+_167298382
|
8.245
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr2_-_85637486
|
8.202
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr1_+_155658881
|
8.182
|
NM_001199849
NM_001199850
NM_001199851
NM_004632
NM_033657
|
DAP3
|
death associated protein 3
|
|
chr3_+_130569368
|
8.118
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr1_-_155948934
|
8.117
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr22_-_42336222
|
8.034
|
NM_001110215
|
CENPM
|
centromere protein M
|
|
chr1_+_174843523
|
8.017
|
NM_001243763
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr5_+_147795459
|
7.830
|
|
FBXO38
|
F-box protein 38
|
|
chrX_-_100872926
|
7.815
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr1_+_155658898
|
7.810
|
|
DAP3
|
death associated protein 3
|
|
chr5_-_133561805
|
7.744
|
NM_002715
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr2_+_87144737
|
7.690
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr12_+_54891522
|
7.688
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr14_-_23426265
|
7.664
|
NM_001166269
NM_001166270
NM_017815
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr4_+_39063851
|
7.652
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr3_+_107241782
|
7.607
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr12_-_121785652
|
7.575
|
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr1_+_155658873
|
7.450
|
|
DAP3
|
death associated protein 3
|
|
chr6_+_64356430
|
7.379
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr20_+_1093905
|
7.362
|
NM_178578
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31)
|
|
chr18_+_32556891
|
7.331
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr3_+_69134057
|
7.274
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr7_+_115863004
|
7.145
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr3_+_172472246
|
7.114
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr17_-_48785016
|
7.114
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr12_+_54891539
|
7.100
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr9_-_15510899
|
7.073
|
NM_001128217
NM_033222
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr2_+_228192227
|
7.062
|
NM_020194
|
MFF
|
mitochondrial fission factor
|
|
chr10_-_98321823
|
7.051
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr9_+_131447364
|
7.035
|
NM_001248000
|
SET
|
SET nuclear oncogene
|
|
chr17_-_38978824
|
7.029
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr6_+_87969619
|
7.022
|
|
ZNF292
|
zinc finger protein 292
|
|
chr13_-_37679800
|
6.991
|
NM_145203
|
CSNK1A1L
|
casein kinase 1, alpha 1-like
|
|
chr4_+_2627158
|
6.946
|
NM_003704
|
FAM193A
|
family with sequence similarity 193, member A
|
|
chr4_+_170581212
|
6.892
|
NM_001243374
|
CLCN3
|
chloride channel 3
|
|
chrX_-_107334749
|
6.874
|
NM_002814
NM_170750
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10
|
|
chr19_-_20607761
|
6.861
|
|
ZNF826P
|
zinc finger protein 826, pseudogene
|
|
chr12_+_54891491
|
6.857
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr6_-_74229094
|
6.819
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr6_+_12717036
|
6.768
|
NM_001242648
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr20_-_45976626
|
6.736
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr6_+_34206567
|
6.556
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr15_-_55563091
|
6.491
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr13_-_50367027
|
6.452
|
NM_002267
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr15_+_100173182
|
6.437
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr2_+_170662044
|
6.424
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr1_-_144520918
|
6.417
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr10_+_112344076
|
6.303
|
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr2_-_152146373
|
6.281
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr20_+_3776385
|
6.266
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr13_+_103459495
|
6.236
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr7_+_141438122
|
6.212
|
NM_003143
|
SSBP1
|
single-stranded DNA binding protein 1
|
|
chr2_+_242255365
|
6.206
|
|
SEPT2
|
septin 2
|
|
chr5_+_67586466
|
6.190
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_-_88119604
|
6.180
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr12_-_80307690
|
6.162
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr14_-_95608054
|
6.149
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr15_+_80352257
|
6.134
|
NM_001242913
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr12_-_102512277
|
6.117
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr6_-_38670829
|
6.114
|
|
GLO1
|
glyoxalase I
|
|
chrX_-_48980070
|
6.000
|
NM_015698
|
GPKOW
|
G patch domain and KOW motifs
|
|
chr12_-_121792011
|
5.995
|
NM_001137559
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr6_-_38670915
|
5.973
|
|
GLO1
|
glyoxalase I
|
|
chr11_-_104827421
|
5.966
|
NM_033306
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr12_-_50616425
|
5.938
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr12_-_122712019
|
5.900
|
NM_019887
NM_138929
|
DIABLO
|
diablo, IAP-binding mitochondrial protein
|
|
chr12_+_28410132
|
5.893
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr13_-_24007822
|
5.875
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr14_-_55650348
|
5.867
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr18_+_33234532
|
5.841
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr8_+_11665394
|
5.782
|
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr8_-_101936521
|
5.776
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr17_+_28707495
|
5.666
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chr10_-_16859408
|
5.620
|
NM_012425
NM_152724
|
RSU1
|
Ras suppressor protein 1
|
|
chr11_-_118900078
|
5.606
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr1_+_148560846
|
5.554
|
NM_173638
|
NBPF15
|
neuroblastoma breakpoint family, member 15
|
|
chr4_-_99851785
|
5.423
|
NM_001130679
NM_001968
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr15_-_55562483
|
5.391
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr6_-_38670904
|
5.353
|
|
GLO1
|
glyoxalase I
|
|
chr5_+_132406080
|
5.339
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr7_+_77469446
|
5.335
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr9_+_75766777
|
5.322
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr9_+_75766794
|
5.314
|
|
ANXA1
|
annexin A1
|
|
chr5_-_145562068
|
5.273
|
|
LARS
|
leucyl-tRNA synthetase
|
|
chr16_-_28437663
|
5.272
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr12_+_56521953
|
5.256
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chrX_-_129272014
|
5.229
|
NM_001130846
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr4_-_72649734
|
5.226
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr7_-_64023331
|
5.223
|
|
ZNF680
|
zinc finger protein 680
|
|
chr7_-_107204705
|
5.186
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr16_+_50321822
|
5.141
|
NM_001114
|
ADCY7
|
adenylate cyclase 7
|
|
chr12_+_56522049
|
5.140
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr12_+_56522043
|
5.106
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr15_+_80352018
|
5.087
|
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr5_+_88185257
|
5.077
|
|
RPS6
|
ribosomal protein S6
|
|
chr3_-_165555252
|
5.065
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr15_+_52201590
|
5.035
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr22_+_21128378
|
5.034
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr2_+_183582667
|
5.031
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr1_-_144520987
|
4.929
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr8_+_110346549
|
4.913
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr5_+_140566655
|
4.908
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chrX_+_119738542
|
4.880
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_-_150738292
|
4.877
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr1_-_151414641
|
4.873
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chrX_+_37639246
|
4.873
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr10_-_75193297
|
4.860
|
NM_001024593
|
ZMYND17
|
zinc finger, MYND-type containing 17
|
|
chr7_+_26400503
|
4.842
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr21_-_47704166
|
4.820
|
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr6_-_79675672
|
4.800
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chrX_-_109683457
|
4.790
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr2_-_88285308
|
4.780
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr6_-_38670882
|
4.767
|
|
GLO1
|
glyoxalase I
|
|
chr2_-_43453724
|
4.764
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr14_+_58906398
|
4.735
|
NM_001244193
|
KIAA0586
|
KIAA0586
|
|
chr6_-_38670871
|
4.732
|
|
GLO1
|
glyoxalase I
|
|
chr1_-_193075186
|
4.709
|
NM_001243399
NM_016066
|
GLRX2
|
glutaredoxin 2
|
|
chr5_+_54398473
|
4.692
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr12_+_12966249
|
4.681
|
NM_016355
NM_201224
|
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47
|
|
chr7_-_64023454
|
4.676
|
NM_001130022
NM_178558
|
ZNF680
|
zinc finger protein 680
|
|
chr5_+_95997871
|
4.665
|
NM_001042440
|
CAST
|
calpastatin
|
|
chr1_-_51887740
|
4.652
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr10_-_16859336
|
4.650
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr12_+_10460416
|
4.627
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr8_-_109260940
|
4.611
|
NM_001568
|
EIF3E
|
eukaryotic translation initiation factor 3, subunit E
|
|
chr2_+_74057969
|
4.593
|
|
STAMBP
|
STAM binding protein
|
|
chr3_-_113897849
|
4.569
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr13_+_53035227
|
4.548
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr5_-_137848517
|
4.545
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr1_+_64088886
|
4.483
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr17_-_47785527
|
4.468
|
|
SLC35B1
|
solute carrier family 35, member B1
|
|
chr10_-_98290309
|
4.459
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr3_-_18480203
|
4.458
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr12_+_14538232
|
4.452
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr2_+_37571752
|
4.446
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr3_-_165555167
|
4.443
|
|
BCHE
|
butyrylcholinesterase
|
|
chr1_+_6105980
|
4.441
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr19_-_8642309
|
4.395
|
NM_012335
|
MYO1F
|
myosin IF
|
|
chr1_-_16134192
|
4.392
|
NM_001089591
|
UQCRHL
|
ubiquinol-cytochrome c reductase hinge protein-like
|
|
chr20_+_32150139
|
4.375
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|